Rajan Gyawali
PhD Candidate in Computer Science
Bioinformatics & Machine Learning Lab · University of Missouri–Columbia
Advisor: Dr. Jianlin Cheng
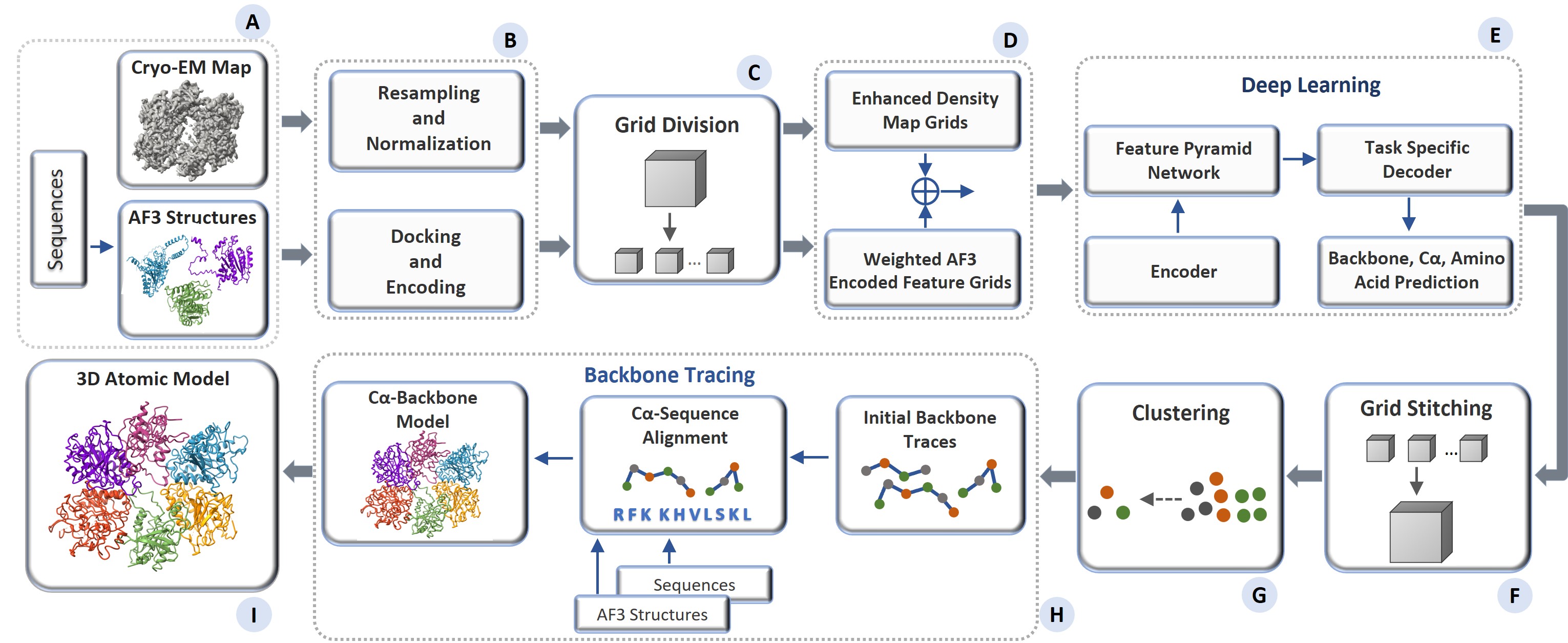
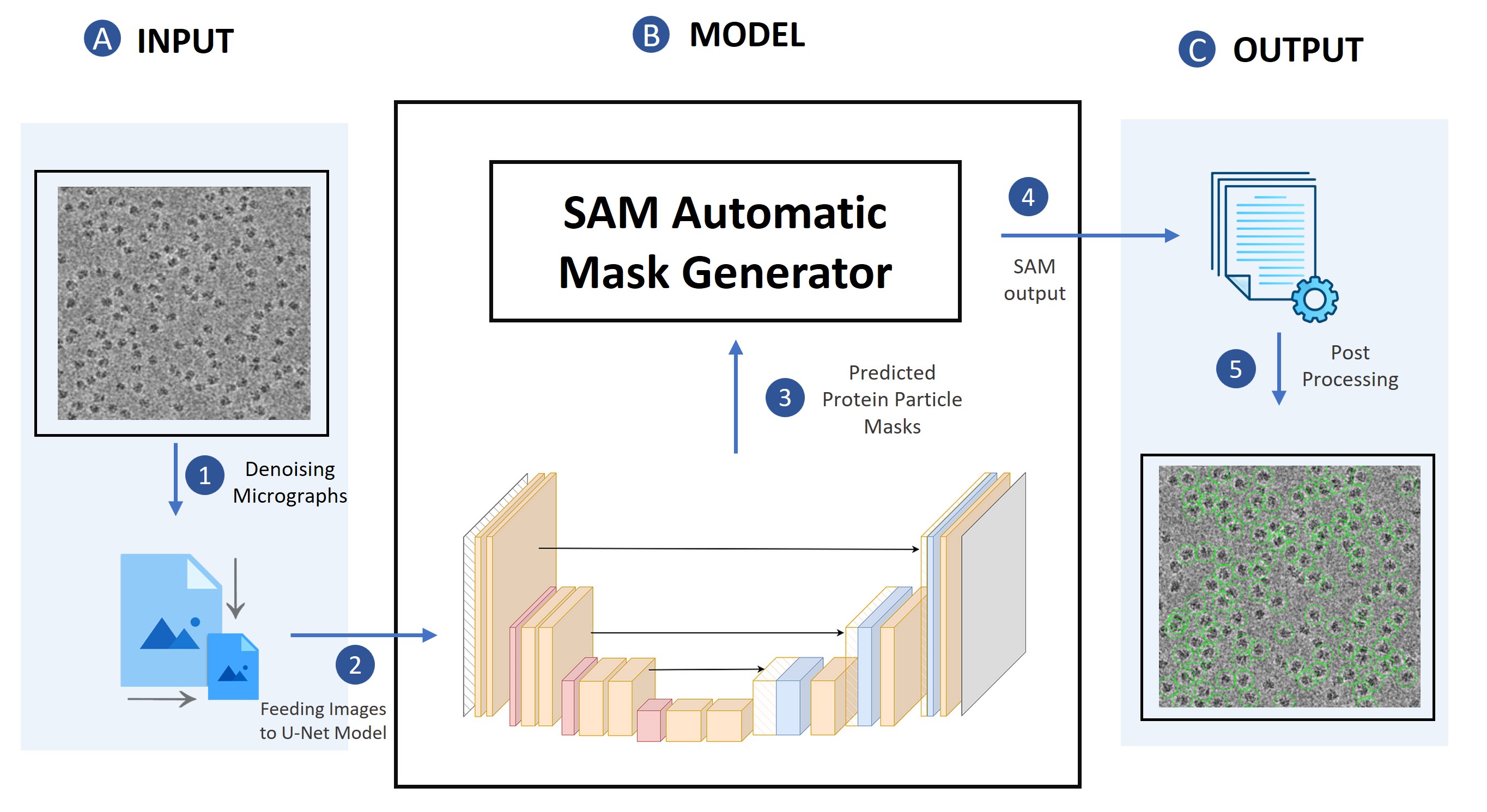
I am a computational biology and machine learning researcher developing deep learning methods at the intersection of AI and structural biology. My doctoral work centers on cryo-electron microscopy (cryo-EM) — designing neural architectures that automate protein particle picking, enable 3D atomic modeling by fusing cryo-EM data with AlphaFold3 predictions, and reduce annotation bottlenecks through few-shot and foundation model-based learning. My methods draw on computer vision, geometric deep learning, and multimodal learning, with applications spanning protein structure determination, virus characterization, and structure-based drug discovery.
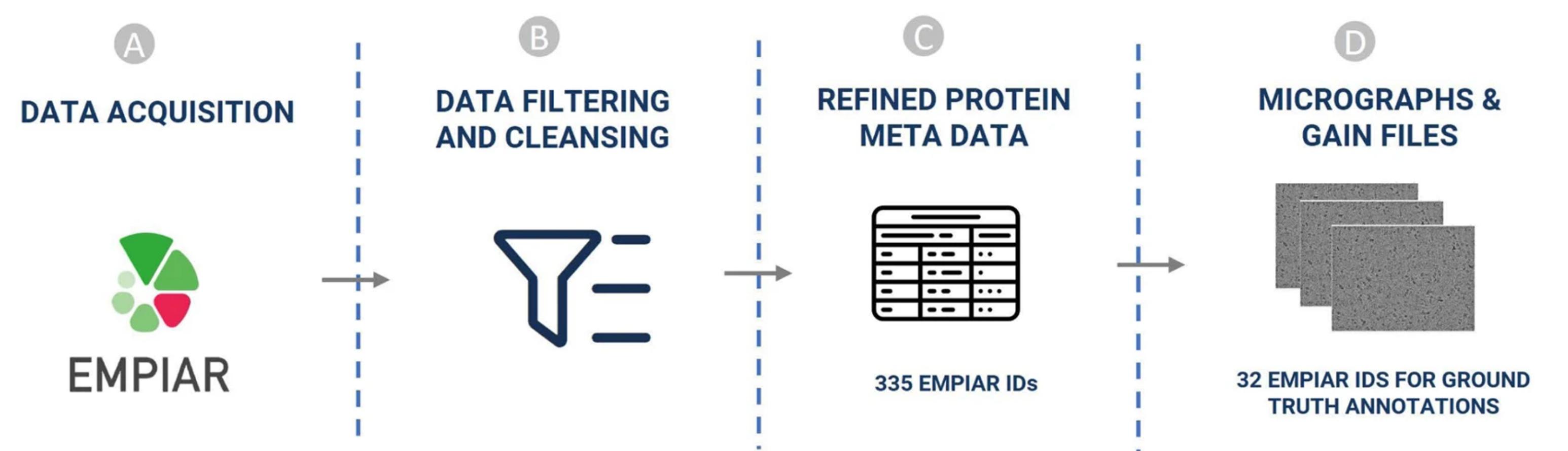
My research is published in leading journals like Communications Chemistry, Scientific Data, Briefings in Bioinformatics and Bioinformatics, with benchmark dataset CryoPPP accumulating 55+ citations and adoption across the global cryo-EM community.